Cores
Core B – Omics and Gene-Editing Core
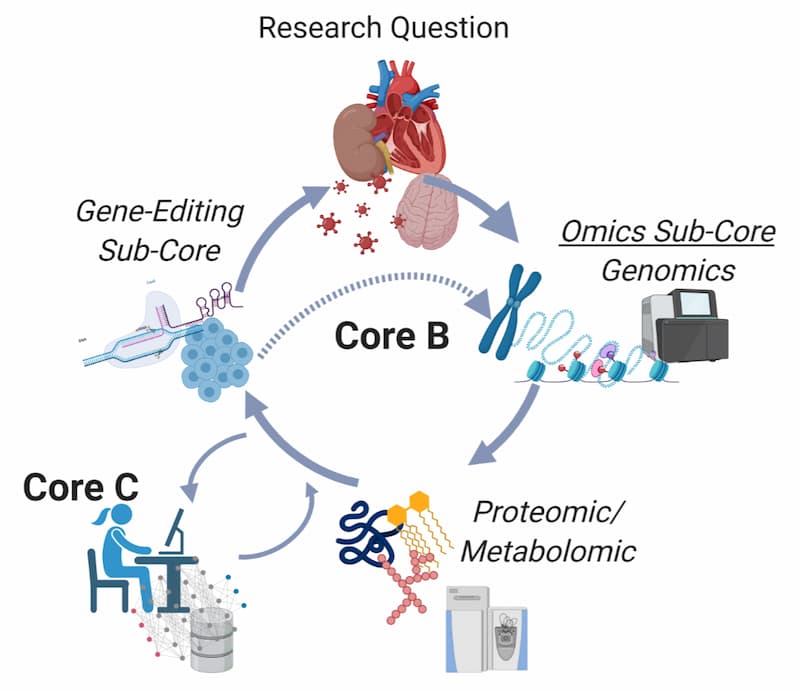 The Omics and Gene-Editing Core provides MCHD investigators access to state-of-the-art omics technologies, including genomics [transcriptome sequencing (RNAseq), whole genome sequencing (WGS), amplicon sequencing (e.g., genotyping by sequencing], 16S/metagenomics, and epigenetic regulation (e.g., methylation)], targeted proteomics and metabolomics, and gene-editing technology. Core B provides support to the centralized UMMC Molecular and Genomics Core Facility (MGCF). The MCHD was central to the recruitment of a world leader, Dr. CB Gurumurthy in gene-editing technologies and the development of animal models. Core B provides support to the newly established Genome Editing Research, Education, and Animal genome Targeting (GREAT) Core.
The Omics and Gene-Editing Core provides MCHD investigators access to state-of-the-art omics technologies, including genomics [transcriptome sequencing (RNAseq), whole genome sequencing (WGS), amplicon sequencing (e.g., genotyping by sequencing], 16S/metagenomics, and epigenetic regulation (e.g., methylation)], targeted proteomics and metabolomics, and gene-editing technology. Core B provides support to the centralized UMMC Molecular and Genomics Core Facility (MGCF). The MCHD was central to the recruitment of a world leader, Dr. CB Gurumurthy in gene-editing technologies and the development of animal models. Core B provides support to the newly established Genome Editing Research, Education, and Animal genome Targeting (GREAT) Core.
Core B provides researchers access to a comprehensive omics data collection pipeline gene-editing and animal model development, and when integrated with Research Computing, Bioinformatics, and Biostatistics Core (Core C) will provide greater depth and insight into biological processes associated with investigator-initiated scientific questions involving the health and disease continuum. Core B in collaboration with the MGCF, will enhance current infrastructure, including new high-throughput sequencing methods such as single cell sequencing, spatial transcriptomics, and long-read sequencing as well as develop new and innovative capabilities, including gene-editing.
The MCHD has partnered with IDeA National Resource for Quantitative Proteomics to provide Discovery Proteomics Capabilities to our researchers. Dr. Allan Tackett serves as a consultant for proteomic applications.
 | Alan Tackett, PhD University of Arkansas for Medical Sciences Deputy Director, Winthrop P. Rockefeller Cancer Institute Distinguished Professor of Biochemistry & Molecular Biology Scharlau Family Endowed Chair in Cancer Research Director, NIH IDeA National Resource for Quantitative Proteomics Director, NIH Center of Biomedical Research Excellence |


